Biotech Data Processing and Analysis
Recent technological advances have led to the generation of vast collections of biotech data. As an example, the European Bioinformatics Institute (EBI) currently stores 20 petabytes of data and back-ups about genes, proteins, and small molecules. This data avalanche has created a strong need for novel mathematical and computational techniques to address challenges related to disease and our environment.
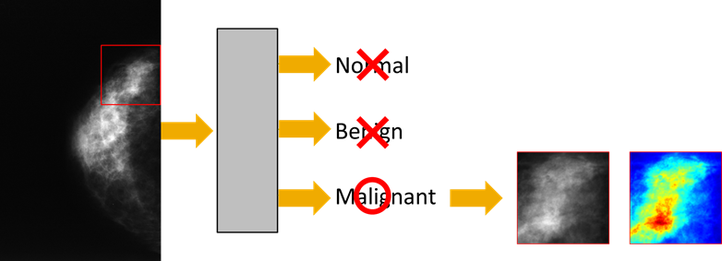
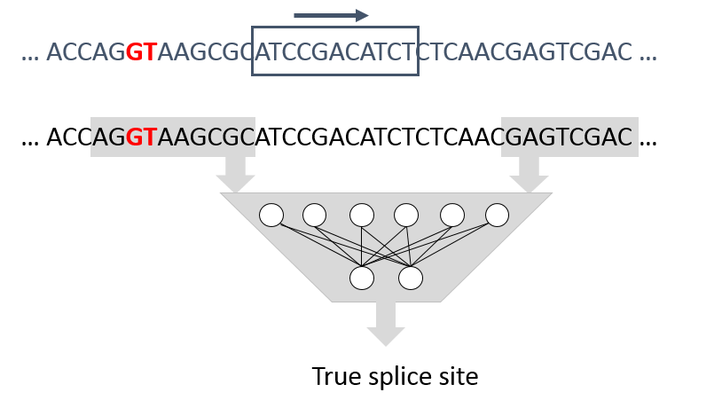
In our research, we explore how multimedia technology can be used to process and analyze biotech data. In that regard, we have two major research objectives. A first objective is to leverage state-of-the-art video compression technology for the compression of genomics data, so to mitigate issues in terms of storage, transportation, and analysis. A second objective is to leverage deep machine learning in the context of several biotech use cases, ranging from genome annotation (e.g., splice site detection in DNA sequences) to tumor detection in medical images. Of particular interest is the application of techniques for natural language understanding to the analysis of genomics data.
Staff
Wesley De Neve, Joni Dambre
Researchers
Tom Paridaens, Jasper Zuallaert, Mijung Kim, Lionel Pigou
Key publications
- Tom Paridaens, Jens Panneel, Wesley De Neve, Peter Lambert and Rik Van de Walle (2016). Leveraging CABAC for no-reference compression of genomic data with random access support. Data Compression Conference (DCC).
- Tom Paridaens, Yves Van Stappen, Wesley De Neve, Peter Lambert and Rik Van de Walle (2014). Towards block-based compression of genomic data with random access functionality. Workshop on Genomic Signal Processing and Statistics. p. 1528-1531
- Tom Paridaens, Wesley De Neve, Peter Lambert and Rik Van de Walle (2014). Genome sequences as media files. 8th International joint conference on biomedical engineering systems and technologies.