BioCAT

Our multidisciplinary team of doctors, engineers, and biomedical experts collaborates to investigate the musculoskeletal system. By combining computational modelling with deep anatomical understanding, we aim to refine diagnostics, and surgical precision and improve positive patient outcomes.
Members
- ZAP: Emmanuel Audenaert (Hip and Biomechanical research), Arne Burssens (Foot and Ankle, WBCT), Wouter Willaert (Anatomy, Surgical Training)
- Postdoc: Hannes Vermue, Matthias Peiffer
- Doctoral researchers: Kate Duquesne, Aline Van Oevelen, Marlies Verleyen, Ide Van den Borre, Roel Huysentruyt, Armin Omidvar Ghaziani, Sara Kowsar, Jing Li, Yukun He, Weilong Xu, Vicky Vandenbossche, Gitte Van Couwenberghe
- Alumni: Leander De Mol
Research projects
1) Motion analysis
Motion analysis is crucial in various fields, including medical diagnosis, biomechanics, and even animation. To acquire kinematic information, optical motion capture is the gold standard. This method tracks reflective markers placed on the skin to infer the underlying 3D movements of the skeleton. Yet, the skin is not rigidly attached to the skeleton, introducing relative skin movements. In computer graphics, this skin movement is considered to be essential to create realistic-looking avatars. However, in rehabilitation sciences, skin motion interferes with the accurate extraction of joint kinematics and kinetics and is therefore considered noise.
In this project, a novel methodology to determine more accurate kinematics is being developed. The skin shift is not treated as noise but is seen as an important source of information. Therefore, methods from clinical biomechanics and computer graphics are blended. The skin geometry deformation is measured and modelled. The kinematics are determined by enforcing consistency between the measured and predicted skin surface.
- researchers: Kate Duquesne, Jan Sijbers, Wim Van Paepegem, Emmanuel Audenaert
2) Medical image segmentation, shape registration and automatic measurements
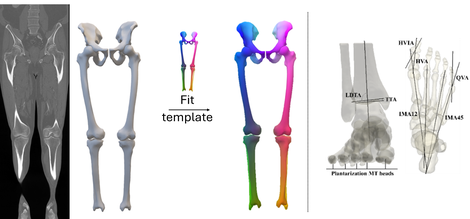
This research initiative is dedicated to leveraging state-of-the-art AI techniques for the precise segmentation of anatomical structures, coupled with shape registration. Our focus is on accurately segmenting bones, cartilage, and menisci, resulting in the rapid generation of 3D models. Particular emphasis is placed on angio CT, Weightbearing CT, and MRI images.
Following segmentation, our process involves registration, where known anatomical landmarks are transferred to new shapes, utilizing developed in-house methods. Additionally, we explore recent advancements in AI-driven mesh processing for enhanced registration accuracy and efficiency. Based on further shape processing and the transferred landmarks, automatic anatomical measurements are performed.
- researchers: Roel Huysentruyt, Ide Van den Borre, Emmanuel Audenaert
Figure 1: Examples of a segmentation and registration procedure (left) and automatic anatomical measurements (right)
3) SHAPE/Genomics
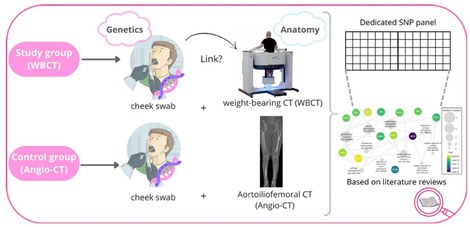
Recent genome-wide association studies have revealed several osteoarthritis (OA) risk loci involve common genetic variations related to musculoskeletal development and morphogenesis. To date, a major shortcoming in this research area is that morphometric phenotyping is based on 2D superposition imaging resulting in high noise levels and lacking applicability. In this interdisciplinary project, DNA is collected from patients undergoing a high-resolution weight-bearing CT scan for medical purposes. To establish the role of shape and biomechanical variance as an independent risk factor in OA, our project provides the opportunity to explore the joint geometry and mechanical phenotype at a population-wide level. Next, subject-specific contact stresses can be computed utilizing a validated discrete element analysis approach. For each configuration, the genetic constitution will be found. Findings will be benchmarked by the identification of high-load-bearing zones in patients having early-stage OA using a large validation cohort of +1000 OA cases.
To create a robust control group, saliva samples, and angio-CT data will be collected from patients visiting the vascular department. A statistical model will be built from the angio-CT data that will be able to predict the skeleton. This model can then be applied to future studies, potentially reducing the need for additional CT scans. The purpose of collecting both CT scans and saliva samples is to establish the relation between SNP genotypes and joint shape variability. However, when we create a well-structured framework utilizing all the available data, a vast amount of research becomes possible for both orthopaedic and vascular patients.
- researchers: Marlies Verleyen, Yukun He, Emmanuel Audenaert
Figure 2: Study protocol SHAPE/Genomics
4) Knee Joint Pressure Calculations For Preoperative Planning
Preoperative planning for knee surgeries plays a vital role in ensuring successful outcomes. Unfortunately, assessing load distribution within the joint, a crucial factor for understanding wear patterns and potential complications remains a challenge in the preoperative setting. Current methods rely on invasive joint pressure measurements, limiting their applicability for preoperative planning. Additionally, traditional methods typically involve analyzing static images, failing to capture the dynamic movement of the knee during activities like walking or running. These limitations can lead to suboptimal surgical plans that do not fully address the specific needs of each patient.
Therefore, this project develops a personalized computational discrete element knee model to non-invasively calculate the pressure on the tibial plateau during gait. Pre-operative simulations using this model can help determine the ideal surgical correction for optimal redistribution of the joint loading.
- researchers: Aline Van Oevelen, Arne Burssens, Amélie Chevalier, Gunther Steenackers, Emmanuel Audenaert
Publications
Contact
Emmanuel Audenaert, PI